QIIME 2 2025.10 is now available!
Hello QIIME 2 Community! 
Our second full release for 2025 is now available! ![]() Huge thanks to everyone for all of their hard work; this is a large release with lots of new and exciting features, bug fixes, and more!
Huge thanks to everyone for all of their hard work; this is a large release with lots of new and exciting features, bug fixes, and more! ![]()
 Important Announcements
Important Announcements 
Job announcement 
- Northern Arizona University is hiring one or more Assistant Professors for the academic year beginning in Fall 2026. We are specifically interested in people who have a background in building and deploying ML/AI methods in support of microbiome research. Have you deployed ML/AI tools in QIIME 2 plugins? That's a plus! Come to NAU and help us lead in microbiome data science! Learn more here.
 Interface Changes coming in QIIME 2 2026.1
Interface Changes coming in QIIME 2 2026.1 
We're planning a broad update to plugins to more consistently refer to metadata, such that many metadata parameters will be renamed to
metadata. This will impact a lot of actions, but will result in more consistent naming so should ultimately make QIIME 2 more convenient for users.--p-n-reads-learnis going to be replaced by--p-n-bases-learnindenoise-singleanddenoise-pairedto match the behavior in dada2
vsearch-global --maxrejectsdefault is going to change
groupis now going to create metadata for the resulting groups, resulting in a new required output
- the
summarize-plusaction will be removed as the currentsummarize-plusaction replaces the existingsummarizeaction.
- Our new experimental taxonomy bar plot, available through the
barplot2command, will be removed as this functionality replaces the currentbarplotcommand. The currentbarplotaction will likely be retained as a newbarplot-oldaction.
- Our new experimental taxonomy bar plot, available through the
Developer Announcements 
Starting in 2026, we’ll update our Python version at most once per year, and only in the April full release (when needed). This gives plugin developers ample time to keep their plugins current and reduces the internal time spend for our team to make these changes each year.
How we decide if we're upgrading our Python version dependency ![]()
After each October release we assess whether a Python update is possible, based on the availability of compatible conda packages for our dependencies, and/or require, based on the end-of-life timeline for our current version.
If an update is planned for April, we start testing immediately so there’s ample time to solve environments and address any API-related test failures.
What this means for developers ![]()
If you maintain a plugin, plan your compatibility work between Oct → Apr (when a Python bump is announced).
No Python bumps will occur in January, July, or October releases.
What this means for users
- Not much. The details of which version of Python QIIME 2 users isn't something users need to be too concerned with.
More details on the schedule and policy can be found here ![]()
 Here are the Highlights of the Release!
Here are the Highlights of the Release! 
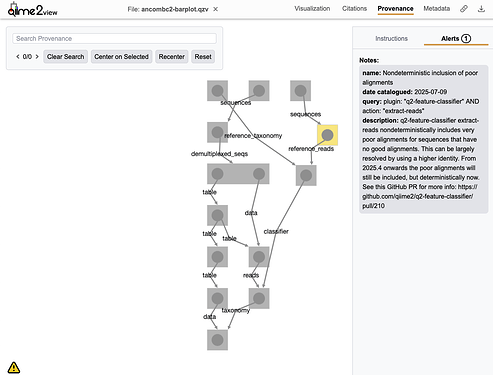
@Oddant1 added provenance-based error reporting to QIIME 2 View. We are now maintaining a database of known issues with plugin versions, so that we can alert users if problems arise (including information on the severity of issues). If you load a Result in QIIME 2 View that is affected by any known issues, the Provenance view will show which nodes are affected and by what issues. Here's an example:
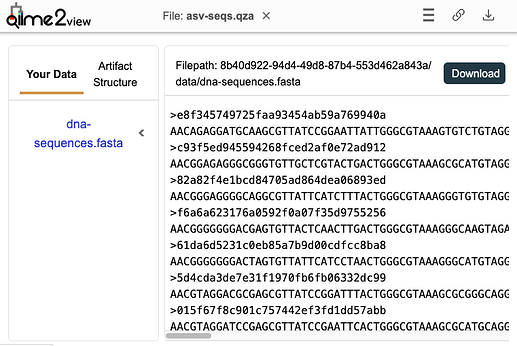
@Oddant1 enabled more informative viewing of QIIME 2 Artifacts (i.e.,
.qzafiles, as opposed to Visualizations or.qzvfiles) in QIIME 2 View. Now, if you load an Artifact, you'll get a preview of and download links for the data they contain. This provides additional transparency on our.qzafile format (remember, it's just a fancy zip file), as well as another easy way to export data from QIIME 2 if you'd like to do that. Here's an example:@jacobs found and fixed a bug in q2view that prevented it from loading URLs with uuids in them here

amplicon
- q2-boots has been added

- q2-fondue has been added

- q2-kmerizer has been added (as a necessary dependency of q2-boots)

- q2-boots has been added
moshpit
- q2-fastp has been added

- q2-fastp has been added
Framework (i.e., rachis) Updates ![]()
The QIIME 2 Framework (sometimes referred to as Q2F) is being re-branded as
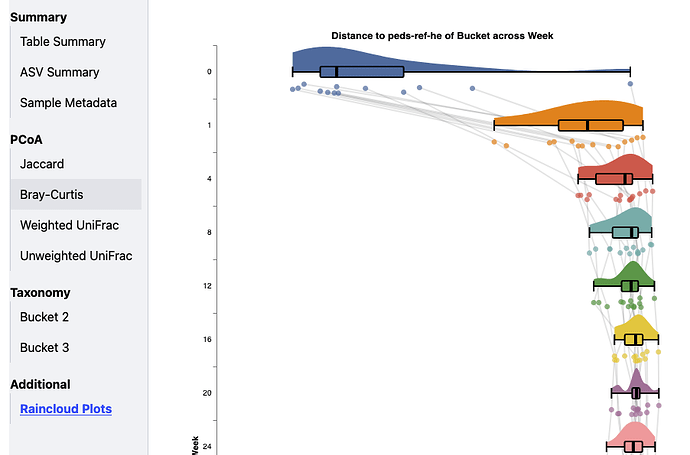
rachis. This won't impact users or developers, but will help us improve clarity in our documentation and is intended to convey generality of Q2F as we explore expanded directions for the project. Learn more in our News post.@ebolyen added a new visualization type called
Report, which enables combining of multiple QIIME 2 Visualizations into a single report that can be shared. This can be accessed using the newqiime tools make-reportcommand, which takes a collection of.qzvfiles as input. It can also be accessed through the Python API. Interact with an example of this on QIIME 2 View here, or see this screenshot. Happy Reporting!
@Oddant1 fixed an issue with recycling output collections when only part of the collection is present. Previously, it would cause an error, now it will only warn then run the action to recreate the collection

@Oddant1 changed the extent of detail in the logs written by actions when run in parallel. These no longer contain
debug-level information, as this was very verbose and made it harder to find relevant information.
@Oddant1 made the behavior of adding multiple copies of the same Result (Results with the same UUID) with different Annotations to the Cache consistent. The Annotations will now be merged in the Cache, and the cached version of the Result will contain all Annotations from all copies

@lizgehret added a new
migrationwarning that can be used for actions or vizualizers that are being moved from one plugin to another. This is a new parameter on theregister_pluginaction, with syntax is as follows:migrated={'to_plugin': 'str', 'from_distro': 'str', 'to_distro': 'str', 'epoch': 'str'}With
to_pluginbeing a required key, and all other keys optional.@lizgehret created the new Archive v7.1 Format, which includes support for cryptographic signing of QIIME 2 Results. This leverages a new Annotation sub-type

Signature, that can be used to self-sign Artifacts or Visualizations, thus allowing confirmation of who created a particular QIIME 2 Result. This provides a path to secure sharing of QIIME 2 Results that contain executable code, such as AI/ML models, that users may want to apply to their private data (for example, taxonomic classifiers or sample classifiers).
- Signatures can be added to an existing QIIME 2 Result (of Archive version 7+) by using the
qiime tools annotation-createcommand. They can subsequently be verified using theqiime tools signature-verifycommand. These actions can also be used via the Python API withartifact.add_annotation(Signature())andresult.verify('signature-name'). - Note that this signing and verifying is only made possible by the existence of GnuPG on your machine, as a GnuPG keypair fingerprint is required for Signature creation, and the existence of the public key connected to an existing Signature on your GnuPG keyring is required for verification. More information on how to install GnuPG and utilize Signatures can be found in our user docs (link to be added here within the next few days).
- Signatures can be added to an existing QIIME 2 Result (of Archive version 7+) by using the
Interface Updates ![]()
- @Oddant1 added a
qiime tools cache-exportbuiltin for exporting artifacts directly from the cache
- @ebolyen added
qiime tools make-reportto generate Reports using the "matryoshka" template (so named because it can be arbitrarily nested, resulting in a hierarchical sidebar containing as many visualizations as you wish to collate). You can see an example here (or in the Report screenshot above).
- @Oddant1 added a
- @ebolyen created a new
matryoshka_templatewhich is for the newmake_reportfunctionality, allowing arbitrarily many visualizations and reports to be collated into a new report.
- @ebolyen created a new
Plugin Updates ![]()
- @jordenrabasco added a new output to each of the denoise methods that records the base transition error rates and added a visualizer (
plot-base-transitions) to display them.
- @jordenrabasco added a new output to each of the denoise methods that records the base transition error rates and added a visualizer (
- @Vinzent_Risch added a
--p-remove-emptyoption to thefilter-samplesaction that allows samples that contain empty fastq files to be omitted in the output.
- @Vinzent_Risch improved the error handling in
filter-samplesif no samples remain after filtering
- @Vinzent_Risch added a
- @Macabe222 changed the plot type of the
mantelvisualizer to a binned variety to fix a bug where very large distance matrices could cause the browser to slow down.
- @Macabe222 changed the plot type of the
- @gregcaporaso added a
baseparameter to Shannon's Diversity Index (theshannon-entropyaction). This enables users to overwrite QIIME 2's default logarithm base of2for this metric to optionally usee(or any other value > 0.0). Providingeas the base will match the Shannon values generated by vegan, scikit-bio, and other tools. At this time, we do not plan to change the default in QIIME 2 for a few reasons:- first, we recommend against direct comparison of alpha diversity values across tools (e.g., vegan versus QIIME 2), as the values are very sensitive to upstream analysis parameters such as those used in quality control and rarefaction;
- second, the relative ranks of Shannon Diversity values for a collection of samples is stable regardless of which base is used for the calculation of diversity for all of those samples - for example, the ranking of Shannon Diversity values will be the same whether they are calculated with vegan (
base=e) or QIIME 2 (base=2); - and finally, changing the default would result in different Shannon Diversity values across versions of QIIME 2, and given the first two points, the juice doesn't seem worth the squeeze.


- @gregcaporaso added a
- @Macabe222 refactored the
tabulate-seqsvisualizer to make the sequence count and length statistics only take into account those records that have DNA sequences.
- @Vinzent_Risch added the ability to provide a random seed to
subsample_ids, improving reproducibility of this Action.
- @Macabe222 refactored the
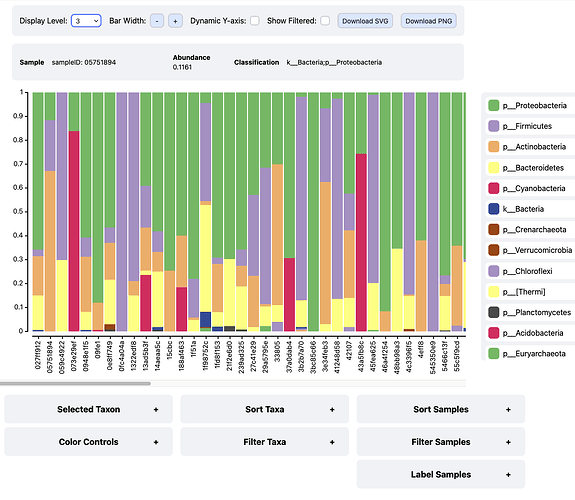
- @colinvwood added a new taxonomic bar plot visualizer (
barplot2) that adds long-requested features such as taxon and sample filtering, expansion of some taxa to lower levels, and to address pain points with thebarplotvisualizer such as poor performance in certain situations. Thebarplot2visualizer should be considered experimental for this release cycle, meaning there is a greater possibility that unexpected/incorrect behavior will occur as it is user tested. In the next releasebarplot2will likely replacebarplot. We're very interested in user feedback onbarplot2- please consider testing it and letting us know what you think, and what other features you'd like to see! This is great chance to impact the future development of this visualizer! Interact with an example here, or just check out this screenshot:
- @colinvwood added a new taxonomic bar plot visualizer (
- @Macabe222 made changes that ensure that paired-end sequencing artifacts allow only forward/reverse read pairs with an equal number of records.

- @Vinzent_Risch migrated the
collate-contigs,collate-genomesandpartition-contigsactions from q2-assembly into q2-types to support more general use across multiple plugins
- @Vinzent_Risch added new collation actions for
GenomeData[Genes]andGenomeData[Proteins]
- @Macabe222 made changes that ensure that paired-end sequencing artifacts allow only forward/reverse read pairs with an equal number of records.
- @Macabe222 unified the subtitle font size for all plots, set a maximum length for all plot titles, and removed grid lines from
heatmap
- @Macabe222 unified the subtitle font size for all plots, set a maximum length for all plot titles, and removed grid lines from
- @SoilRotifer updated the action
get-pr2-data:- Removed PR2 reference database
4.14.0, as this database was never meant to be fetched as it only contains 8 taxonomic ranks. This action was intended to fetch recent databases that contain 9 taxonomic ranks. - PR2 reference database
5.1.0can now be downloaded. - code for this action was refactored to enable more streamlined unit testing.
- Removed PR2 reference database
- @SoilRotifer updated the action
get-midori2-data:- users can now fetch the recent MIDORI Reference 2 sequence & taxonomy data:
GenBank266_2025-04-24andGenBank267_2025-06-19.
- users can now fetch the recent MIDORI Reference 2 sequence & taxonomy data:
- @SoilRotifer added the action
get-eukaryome-data. Users can now download reference sequences and reference taxonomy from the Eukaryome reference database. Options include:- the ability to download separate SSU, LSU, and ITS sequences
- the entire region that contain these genes , i.e. "longread".
- can download all available data simultaneously for a given database version.
- @SoilRotifer added migration warnings for the following actions, which are tentatively slated for the
2026.1release of QIIME 2:evaluate-cross-validatewill be migrated to theq2-feature-classifierplugin.evaluate-fit-classifierwill be migrated to theq2-feature-classifierplugin.trim-alignmentwill be migrated to theq2-alignmentplugin.
- @SoilRotifer updated the action
Community Plugin Updates ![]()
- plugin template
- @cduvallet fixed a small

- @cduvallet fixed a small
Docker Container Updates ![]()